Spooky2 is being continually refined for the betterment of mankind.
Changes since the previous release:
Improved: Calculation method of genome frequencies. It is explained at the end of this release announcement.
Some people have noted how some DNA frequencies worked, whilst others seemed to have no results. The cause of this phenomenon has been identified, and the method of frequency derivation has been refined. In this paper, the new process of calculating DNA, RNA, and mRNA frequencies is explained.
The new factors are incorporated in this release of Spooky2.
RNA to Hz and mRNA to Hz factors have been added to Spooky2:

The database holds the genome size and type:
- Bx x is the number of Base Pairs in a genome.
- BCx x is the number of Base Pairs in a Circular genome. BLx x is the number of Base Pairs in a Linear genome.
- R defines an RNA entry. Example: BLR29900, BCR200
- m defines an mRNA entry. Examples: BLm29900, BCm3560
Improved: Program loading speed. Larger databases take longer to load. The Spooky2 program loading code has been optimized to ensure even very large databases load promptly. The MicroGen software has also had this improvement.
New: Support for GX Pro. The older GX has a frequency resolution of 5 decimal places up to 40 kHz, and 2 decimal places above 40 kHz. GX Pro has a frequency resolution of 8 decimal places for the entire frequency range.
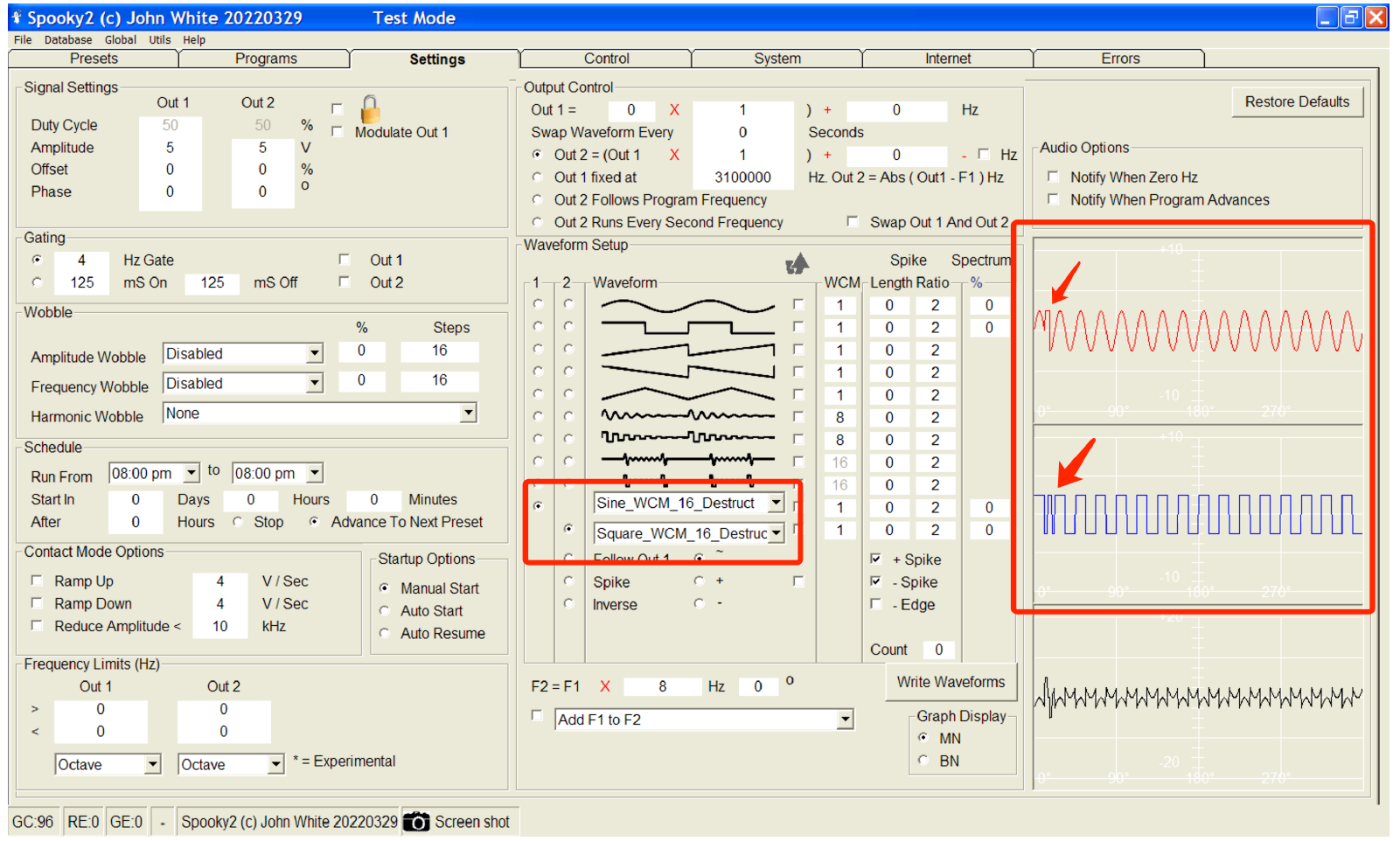
New: Sine and Square waveforms for destructing. These waveforms have a reversing spike on every 16th wave cycle. At the point where the target reaches maximum resonance kinetic energy, it hits “the brick wall”, causing the greatest stress to the genome or target pathogen.
New: Spooky2 software is complicated to use. We have introduced a Normal Mode with simplified functionality for new users. By default, Spooky2 will start in Normal mode. The top menu toggles between Normal and Advanced modes.

Fixed: Slave generator beat frequency bug.
These people contributed their time to help make this release of Spooky2 special:
Bryan Yamamoto
Ed Von
John Robert Grayson
Walt Dod
Derivation of Genome Frequencies Rev. 2
By John White
problem Clinic NZ Ltd.
https://www.spooky2.com
March 29th, 2022
The following document is gifted to humanity. There is no copyright. The information herein may be freely shared by everybody. Additionally, no patents have been, nor ever will be, applied.
Introduction
This paper revises the Derivation of DNA Frequencies documents released May 10th, 2020.
Some people have noted how some DNA frequencies worked, whilst others seemed to have no results. The cause of this phenomenon has been identified, and the method of frequency derivation has been refined. In this paper, the new process of calculating DNA, RNA, and mRNA frequencies is explained.
Historical Background
As far back as the 1930s, a brilliant scientist named Royal Rife designed electrical frequency devices which disabled pathogens and relieved serious illnesses such as problem and pneumonia. He discovered that each pathogen had a specific disabling frequency.
To discover the frequencies, Rife designed immensely powerful microscopes. Through meticulous observation, Rife could identify which frequencies disabled the pathogens. He found that the frequencies must be precise. Other frequencies had no effect on the pathogens.
People often wonder why modern frequency therapeutical devices are not as effective as these very first machines. The reason is very sad. Rife’s microscopes and electrosolution devices did not survive the test of time. Most of them were damaged beyond repair. Technical knowledge has been lost, and we no longer have the ability to watch live pathogens at such high magnifications. Without the correct frequencies, modern machines cannot match the breathtaking performance of Royal Rife’s antique machines.
Rife’s life-long experiments proved that pathogens can be quickly destroyed through the use of resonant frequencies. Rife used the term “absolute coordinative resonance” to describe how his frequencies worked.
Resonance
Playground swings are a very simple example of resonance. Gentle pushing at the right moment (frequency) causes a child to swing higher and higher. The energy of each push adds kinetic energy to the swing. If the pushes are not timed correctly, resonance is lost and the swing energy is reduced. You also get a grumpy child.

Radio receivers also use resonance. When a radio is correctly tuned, it resonates with the radio station’s transmission frequency.
DNA and RNA are both genomes. Genomes hold the complete set of genetic information of an organism, providing all of the information the organism requires to function. One of the main functions of DNA is protein production.
Resonant frequencies can prevent genomes from creating proteins and replicating, thus disrupting the life functions of the cell. If the cell is a pathogen, it would become non-viable, and an easy target for our innate immune system.
The DNA and RNA Antenna
Genomes form biological monofilar and bifilar helical antennae tuned to specific frequencies. Applying these frequencies causes them to resonate.

This idea is not new. A patent was released almost 15 years ago (Charlene A. Boehm, METHODS FOR DETERMINING THERAPEUTIC RESONANT FREQUENCIES, US 7,280,874 B2, Oct. 9, 2007) describes how to derive DNA resonant frequencies from the axial length of the DNA strand.
The resonant frequency of normal-mode helical antennae is determined by the radial length of DNA, not the axial length. This is the length of the sugar-phosphate ‘backbone’ shown in red below. If the radial length of the genome sugar-phosphate ‘backbone’ is the same as the applied wavelength, the conditions for resonance are met.
The antenna can be either a closed-loop (circular genome) or open-ended (linear genome).
RNA vs DNA
When calculating the resonant frequencies of genomes, structural differences must be considered. RNA has an extra OH group on the sugar ‘backbone’, so the twists are tighter. The effective helix radius of RNA is also different from that of DNA. These factors cause RNA to resonate at a different frequency to DNA, even if they have the same base pair count.

The Maths
Genome resonating frequencies can be calculated using this formula:
Frequency = EMF propagation speed / effective genome length.
The EMF propagation speed is the speed of electrical frequencies traveling through a medium. The medium of DNA and most RNA is nucleoplasm.
Genome resonance calculations must consider the effective genome length. There are two basic configurations for genomes. Linear genomes are the most common.
Calculations for this genome type require a base pair to be subtracted when calculating the total length. This is the effective base pair count (bn).

The above graphic represents a linear double-strand DNA genome with 3 base pairs. There are only 2 gaps between them. The total genome length is (base count – 1) x (distance between base pairs). Circular genomes do not require a base pair to be subtracted.
Genome resonance occurs when the wavelength of the applied EMF signal is an octave of the genome length. Octaves are factors of 2 raised to a power. Examples of octaves are 2, 4, 8, 16, and 32.
EMF travels at the speed of light through a vacuum: 2.99792458E+8 meters per second. Most genomes are immersed in the nucleoplasm, where the EMF propagation speed (vn) is slower: 1.23706749665013E+08 m/s.
Radial genome length can be calculated using this formula:

mRNA
The differing dimensions between DNA and RNA result in different resonant frequencies, but there is another factor that must be considered. mRNA moves to the cytoplasm where proteins are made. The EMF speed through the cytoplasm is 1.15164716490445E+08 meters per second. Therefore, mRNA resonant frequencies are not the same as DNA or ‘regular’ RNA, even if the effective base pair counts are the same.
Working Examples
Working examples of DNA and RNA frequency derivation is shown in this table:

The resultant frequencies are very high but can be reduced using a sub-harmonic of choice. Most commonly, sub-octaves are used, to align with standard antennae theory and practice. These are the inverse of two raised to an integer power, ie, 2, 4, 8, 16, 32, etc.
Conclusion
Pathogens have 3 main types of genomes: DNA, RNA, and mRNA. Each will have different resonant frequencies, even if their respective base pair counts are the same.
By applying radio principles, it is possible to calculate accurate resonant frequencies of target genomes. Sub-harmonics of this frequency can then be applied, disrupting the life functions of pathogens.
General Notes
People have asked what makes our genome databases and presets superior to DNA frequencies from other sources. Here are some of the reasons:
• Genome strands act as a spiral antenna. Spiral antenna frequencies are calculated using radial lengths. For this reason, radial lengths are used in our formulae.
• Genome strands are constantly moving, expanding, and contracting like a spring. Unlike linear length, the radial length remains constant.
• The correct value of permittivity is used.
• The significant structural differences between DNA, RNA, and mRNA are taken into consideration.
• Fundamental frequencies are calculated, allowing people to unleash the maximum potential of their frequency equipment.
• Viruses and other pathogens continually mutate. Spooky2 DNA databases are updated regularly to catch new strains and omit genome strains that have gone extinct and are no longer relevant.
• Spooky2 DNA databases and presets are free.
• Spooky2 DNA frequencies can be used on any device. They are not machine-specific.
• Spooky2 DNA frequencies are harvested from multiple sources and are updated regularly to catch virus mutations and genome corrections.
• The Spooky2 software calculates frequencies on-the-fly for razor-sharp accuracy and maximum resolution.
Credits
I would like to thank the moderators of the Spooky2 forum for assisting in the creation of this document.
I would also like to thank Charlene Boehm. Her pioneering attempt to match DNA length to frequencies provided the impetus to formulate the correct frequencies.
Disclaimer
problem Clinic NZ Ltd, Clean Technologies, John White, Echo Lee, supporting staff, and crew (hereafter referred to as Team Spooky2) ARE NOT RESPONSIBLE for any damage or injuries of any sort or form that may be sustained by any person or persons, any animal, or to any equipment or any other thing or things. The Spooky2 DNA Frequencies have not been approved for use by any governmental or medical agency or inspection service. No medical claims are made for, nor implied by Team Spooky2.
You are advised to always consult with your physician or other health care professional if you have, or think you might have a health problem.
